Innovative nanosensor method for monitoring viral vector quality in gene therapy
In recent years, the field of genetic engineering has made tremendous strides, bringing us closer to a future where we can alter genes within living organisms. This could pave the way for gene therapies that may revolutionize the medical landscape. Currently, one of the most promising avenues for such therapies involves using the molecular systems found in viruses as delivery mechanisms.
Among these, adeno-associated virus (AAV) vectors have recently stood out due to their potential for developing nucleic acid-based vaccines, with applications in diseases such as COVID-19. However, one challenge that arises during AAV production is that some vectors may carry incomplete versions of the desired genetic material, while others might be entirely empty. Such defective vectors can cause unforeseen reactions, highlighting the critical need for improved quality control measures in vector manufacturing.
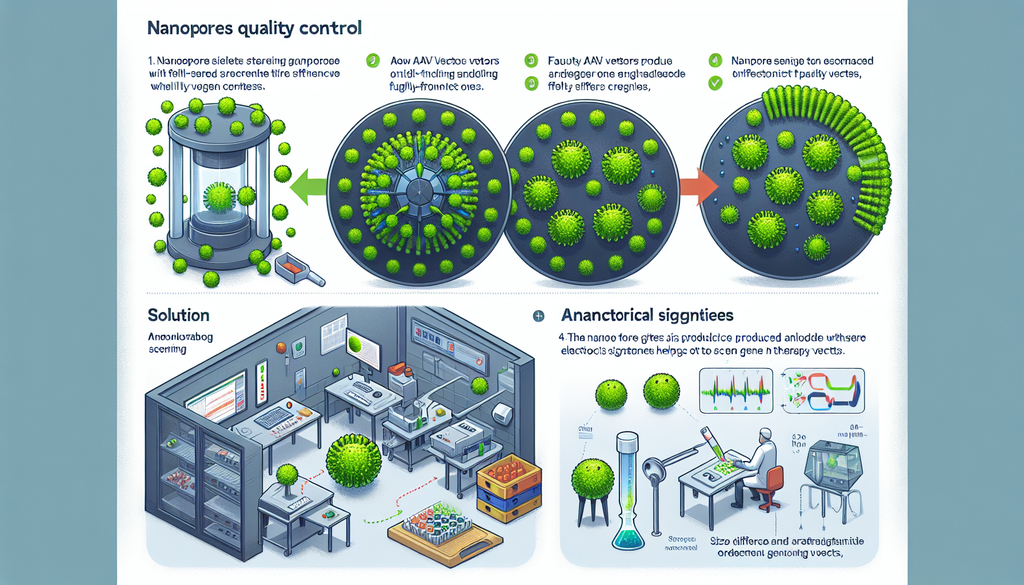
To address this issue, a group of Japanese researchers introduced a groundbreaking nanosensing method that enables the analysis of viral vector properties. Their novel technique was presented in a paper published on June 5, 2024, in the journal ACS Nano. The research team comprises a multidisciplinary group, including Associate Professor Makusu Tsutsui and Professor Tomoji Kawai from Osaka University’s Institute of Scientific and Industrial Research, Lecturer Akihide Arima and Professor Yoshinobu Baba from Nagoya University, and several scientists from the Institute of Medical Science at The University of Tokyo, such as Project Researcher Mikako Wada, Assistant Professor Yuji Tsunekawa, and Professor Takashi Okada.
The researchers designed a method that measures the ionic current through a nanopore in a solution containing AAV particles, applying an electric voltage. When unimpeded, the ionic current remains stable, but each time a viral particle passes through the nanopore, it briefly obstructs the flow of ions. This interruption registers as a distinct spike or fluctuation in the current.
What’s fascinating is that the researchers discovered a way to differentiate between full-genome vectors and defective ones. Since AAV vectors containing complete genomes are slightly larger and heavier, they create unique signatures in the ionic current compared to empty or partially filled vectors. The team confirmed this through a combination of laboratory experiments, simulations, and theoretical modeling. "By optimizing the nanopore sensor’s design, we were able to detect sub-nanometer-sized differences between the viral vectors for the first time," Tsutsui elaborates.
This method provides an efficient and cost-effective means for quality control in the production of AAV vectors. Previously, complex tools such as mass photometry, transmission electron microscopy, and ultracentrifugation were necessary for such tasks. “Our findings could fundamentally change the field by offering a tool for producing AAV vectors with extremely high precision, crucial for their safe and effective use in gene therapy,” says Tsunekawa. He adds that this could significantly improve production and purification processes for AAV vectors.
Beyond just AAV vectors, this technology may also be applicable to studying other viral vectors. It represents a potential breakthrough, fostering new developments in gene therapy and expanding our understanding of viruses. Ensuring the quality of vectors used clinically might even reduce necessary dosages, thereby minimizing potential side effects for patients.
There is hope that these advancements will bring us closer to effective treatments for genetic disorders and difficult-to-treat diseases such as cancer.
Journal
ACS Nano
DOI
10.1021/acsnano.4c01888
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Identifying Viral Vector Characteristics by Nanopore Sensing
Article Publication Date
5-Jun-2024